|
Table of Contents
Mining Globins FamilySetting up the experiment dataset
#shortFamilyName long-family-description pdb1 pdb2 ... where pdb1, pdb2 are pdb names with chain identifiers (as for example, given in SCOP). You can download the globins and kinases dataset file as it was prepared by Aung, et.al., 2004 and place it into the Parameters folder. Pre-processing the experiment dataset
>> globals_set();
>> [ptns,fams]=data_prepare_experiment('experimentFile','families.globins.kinases.txt')
1- 10 a.1.1.2 - (Globins)
11- 28 a.1.1.* - (Globin-like)
29- 38 d.144.1.7 - (Ser/Thr Kinases)
39-227 _except_a.1.1.* - (except Globins)
228-424 _except_d.144.1.7 - (except Ser/Thr Kinases)
ptns =
1x278 struct array with fields:
pdb
features_cp_normalized
fams =
1x5 struct array with fields:
name
title
ind
members
range
numMembers
For each family, Mining for significant sites
After ensuring that all pdb files are present, the >> rep=mine_family_represent('globins', 'ptns',ptns([1:10]), 'rand',ptns([39:227]),'recache',1)
rep =
feats: [1296x14 double]
scores: [1296x1 double]
borderDs: [1296x1 double]
threshold: 0.2560
Here, we gave globin-like proteins (1 through 10) as the family to be mined, and the rest of the proteins in the dataset as the outgroup. The return value of Examining and displaying the sites
The returning value >> signatures_show('1irda', 'familyName','globins', 'numHits',1, 'pretty',1);
1: 1 -->208. score=0.000 dist=0.035 count=1
residues: PHE43 HIS58 HIS87 TYR42 PHE46 LEU91 LEU29 HIS45 VAL62 LEU86 PHE33 LEU83 VAL93
atoms: PHE43(N,CB,CG,CD1,CA,CE1,CZ,CD2,CE2) HIS58(CB,CG,ND1,CD2,CE1,NE2,O,CA) HIS87(CA,CB,CG,ND1,CD2,CE1,NE2) TYR42(CB,CG,CA,CD1,C,O) PHE46(CE1,CZ,CD1,CE2) LEU91(CB,CG,CD1,CD2) LEU29(CD1,CD2) HIS45(CE1,NE2) VAL62(CB,CG2) LEU86(CG,CD2) PHE33(CZ) LEU83(CD1) VAL93(CG2)
drawing the protein...
drawing the local sites found...
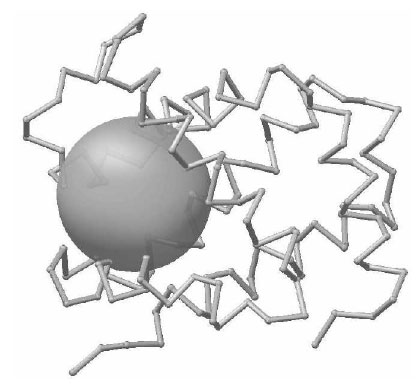
In the Matlab window, you can use click&drag the mouse to rotate the protein. To turn on the labelling of the residues, pass the parameters 'label',1. The figure above shows the top representative critical point with its spherical neighborhood, perfectly allocated at the heme-binding pocket of the protein 1irdA – human hemoglogin alpha subunit. This metal binding pocket is highly conserved in the Globins family and its presence is critical for the function of the protein. The Histidine residues 58 and 87 responsible for binding Iron atom are contained within this spatial neighborhood. The residues within the representative site are given in the output, along with the detailed atom list.
It is also possible to see the rest of the features mapped on to the protein space (use |